This year’s Nobel Prize in Chemistry is awarded to David Baker (University of Washington, Seattle, USA), Demis Hassabis, and John Jumper (Google DeepMind, UK) for their work on developing Rosetta (Baker) and AlphaFold (Hassabis and Jumper). These open-source software tools enable researchers worldwide to go from an amino acid sequence to a 3D structure of a protein with just one mouse click.
‘It used to take us 3 to 4 years to determine the structure of a specific protein, but we can now get an accurate prediction of that structure almost immediately’, explains Prof. Yann Sterckx, associated with the Laboratory of Medical Biochemistry led by Prof. Ingrid De Meester at the University of Antwerp.
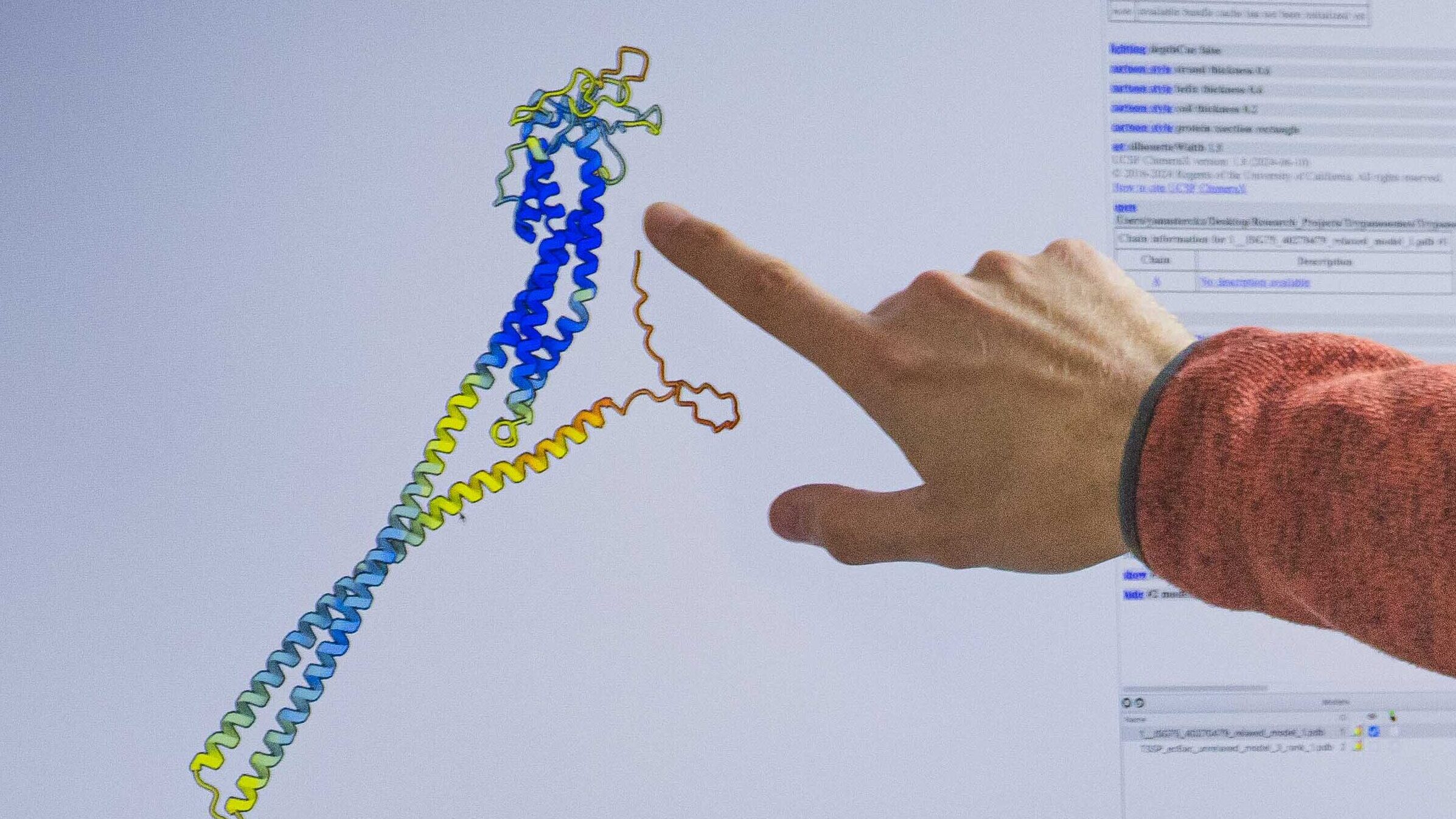
As a structural biologist, Prof. Yann Sterckx has been working with AlphaFold software since its launch in 2021. ‘It has accelerated our work’, he explains. It’s a tool used daily in laboratories all over the world.’
“It used to take us 3 to 4 years to determine
the structure of a specific protein, but we can now
get an accurate prediction of that structure almost immediately.“
Software developed using machine learning is only as good as the data available for it to learn from, and this is no different for AlphaFold. Since the 1970s, scientists have been pooling experimental protein structures in a central database, the Protein Data Bank (PDB). Based on the more than 225,000 protein structures now in that database, AlphaFold can predict a 3D structure from a sequence. ‘We provide the software with a sequence of amino acids’, explains Prof. Yann Sterckx. ‘The software, installed on a powerful computer (e.g., the Flemish supercomputer), performs the heavy calculations, and soon after, we have a workable prediction of the protein structure.’ A color code, among other things, indicates the accuracy of the prediction. If the structure is blue, you can be sure the prediction is correct. If the protein structure is orange, the prediction should be taken with caution. ‘It is still up to the scientist to make the correct assessment’, explains Prof. Yann Sterckx.


Proteins are the workhorses of our body, and of all living organisms. But they are also involved in many diseases. Human proteases (Prof. Ingrid De Meester’s research focus) play a role in cancer and inflammatory diseases. Proteins also play an important role in infectious diseases. Protozoan parasites, such as those causing malaria and leishmaniasis, also rely on proteins to cause infection. ‘In our lab, we conduct research on infectious diseases, specifically on the interaction between the parasite and the host.’ Parasites are constantly seeking clever ways to evade our immune system. ‘I study how the parasite uses its proteins to attach to host molecules, which can also be proteins’, explains Prof. Yann Sterckx. ‘Knowing the protein structure is crucial to better understanding the infection process.’
Also interested in UAntwerp Biochemistry study program

